New Cas9 model maps DNA cutting behaviour for the first time
Researchers from the TU Delft have come up with a physical-based model that establishes a quantitative framework on how gene-editing with CRISPR-Cas9 works, and allows them to predict where, with what probability, and why targeting errors (off-targets) occur. This research, which has been published in Nature Communications, gives us a first detailed physical understanding of the mechanism behind the most important gene editing platform of today.
The discovery of the CRISPR-Cas9 protein has greatly simplified gene editing, and raised hopes to find a cure to many hereditary diseases. However, routine and safe use of this technique in human therapeutics requires extreme precision and predictability of any off-target effects (errors). A research team lead by Martin Depken at TU Delft’s department of Bionanoscience has now demonstrated a new, physical-based model that greatly improves on existing models: not only does the model predict where the DNA is likely to be cut, but also with what probability this will happen.
Physics-based approach to understand Cas9 gene-editing
A great limitation of current bioinformatics models for gene editing lies in the fact that they are binary in nature: they classify targets on the genome as either likely or unlikely to be cut. These models focus only on very high-probability targeting errors (off-targets), and will miss the many lower probability off-targets that together could amount to the majority of editing errors in the genome. Now, the new physical model which the researchers created takes into account both high-probability and low-probability off-targets; it can be used to physically characterize any Cas9 variant and predict the probability that any site will be cleaved.
Martin Depken explains his lab’s new physics-based approach: “In gene editing, you want to maximize the probability of cutting at the intended site, while minimising the amount of cutting in the rest of the genome. It is therefore crucial to understand cutting in terms of probabilities. Drawing from experiments in single-molecule physics and structural data, we created a model that can do this. We changed the way in which to describe the gene editing from a binary choice to a complete probabilistic picture.”
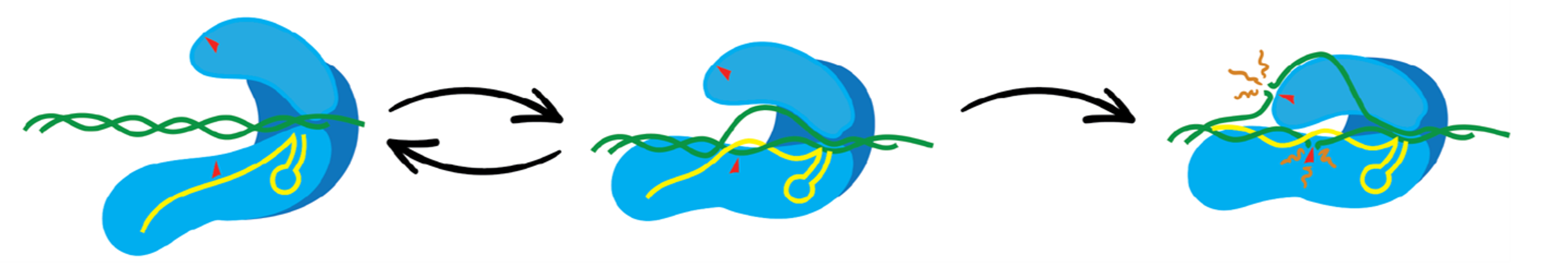
Improving accuracy in gene editing
By giving physical insights into why off-targets (errors) occur, this research also marks an important next step towards a more rational way of engineering new gene-editing platforms, and for characterizing, evaluating, and comparing existing ones. With their probabilistic description of gene-editing, the researchers also hope to help improve the risk assessment in genome editing by accounting for all possible off-targets.
“Together with our experimental collaborators at University of Texas at Austin, we’ve now benchmarked Cas9 with our model,” says Depken. “Our next goal is to do the same with other high-precision gene-editing platforms to understand how and why they differ. With this we hope to reveal the path to even higher precision in gene editing.”
