Unravelling how DNA is looped with DelftBlue and experiments
Proteins are the molecules that carry out essential functions in our cells, such as compacting long strands of DNA into a tiny structure in the cell nucleus. Experimental biophysicist Roman Barth wanted to understand this process down to the molecular level. Thanks to the DelftBlue supercomputer, he was able to compress several years of experiments into a single year.
Knowing the sequence of amino acids that a protein is made of doesn’t tell you its three-dimensional shape. And knowing its shape doesn’t tell you its function and how it interacts with other molecules when performing that function.This is the challenge Roman Barth faced during his PhD at the Cees Dekker Lab in the Bionanoscience Department.
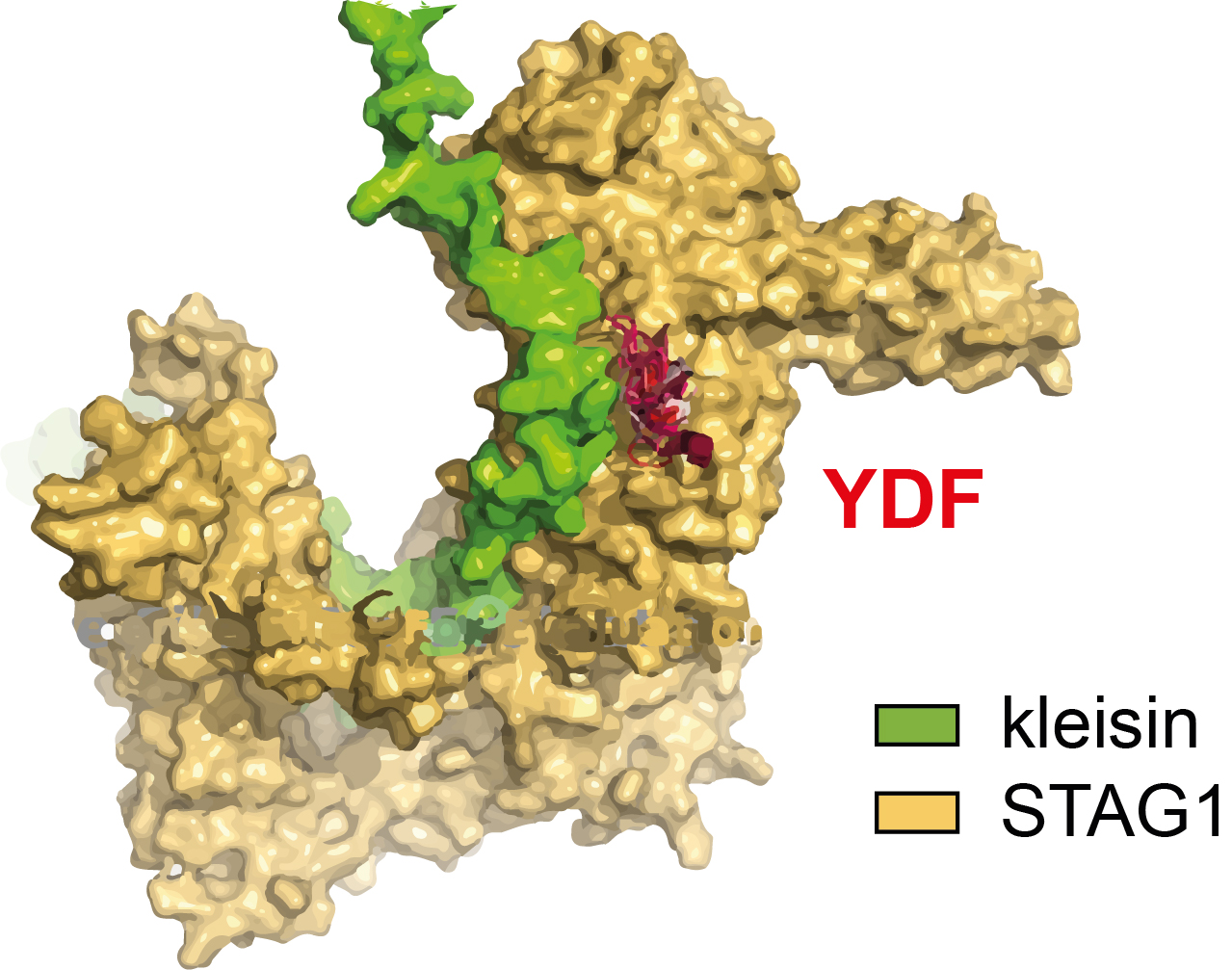
“My goal was to unravel the interaction between two proteins known to be involved in the compaction of DNA via a process called DNA loop extrusion,” he says (see video ). “The first protein, cohesin, will loop DNA continuously if unstopped. The second protein, CTCF, acts as a stop sign to cohesin. That is very important for the cell as DNA, for example, needs to be accessible when read, expressed or maintained.”
The interactions between proteins can’t be calculated from their amino acid sequences using first principles.
AlphaFold
The interaction between the two proteins depends on their shape, the parts that are in close proximity, and the forces that then occur between groups of atoms. “These can’t yet be calculated from their amino acid sequences using first principles,” Roman says. “And while CTCF is a relatively short protein, cohesin is a complex of five subunits which it can exchange – allowing for many configurations and many more interactions.”
At the start of the project in 2023, it looked like a brute-force experimental approach was the only way to go. But then AlphaFold came along. “AlphaFold is an algorithm, based on artificial intelligence, that can predict the structure of proteins from their sequence,” Roman says. “It completely changed our approach. We could now make predictions about what our proteins would look like, and what may happen to them when they meet. Instead of experimentally testing all possible configurations of CTCF and cohesin, we could ask the computer to predict likely interactions and test only those.”
Project storage allows multiple users from various departments to share resource-intensive installations on DelftBlue.
Powerful computer
AlphaFold did come with the new challenge of requiring a powerful computer to run on – a lot of disk space to store the 5 Tb database it browses and a lot of computing power to run the algorithm. “It is almost impossible to run AlphaFold on a laptop or desktop and we lacked the expertise to build our own cluster. So, our second lucky break was that DelftBlue came online.”
It also turned out that another researcher, Marcel van den Broek from Biotechnology, had already installed AlphaFold on DelftBlue. Rather than having every user install their own version, the DelftBlue administrative team setup what is now called project storage. “It allows multiple users, from various departments, to share such resource-intensive installations. Marcel and I were the first to make use of this feature for AlphaFold.”
Cluster newbie
Roman was still pretty much new to using a computational cluster when he started with DelftBlue, but he quickly found his way. “DelftBlue has an intuitive interface, great documentation, and a very knowledgeable and helpful support team,” he says. “You can basically figure it out from there.” He avoided running into the 5-day runtime limit for jobs submitted by breaking up the CTCF protein into smaller fragments. “Still, it would be nice for DelftBlue to have even bigger GPUs or to be able to use multiple GPUs at the same time, especially for tasks involving artificial intelligence.”
One thing he really appreciated was that the DelftBlue administrators were very open to users trying out new things. “Their attitude of ‘seems to be useful, we’ll give you the resources you need’ has been very helpful,” Roman says. And when he ventured into new DelftBlue territory, such as when he submitted hundreds of jobs at once, the administrators were just as curious about how the cluster’s performance. “Turns out the job allocation scheduler handled it well.”
Testing everything purely experimentally would have taken us at least three times as long.
Two likely interactions
Using AlphaFold and DelftBlue, Roman was able to pre-screen hundreds of different combinations. In this way, two likely interactions were identified. These were validated in laboratory experiments in which he looked for how these two fragments of the CTCF protein affected the DNA looping process. “Even though any such fragment can nowadays be quite easily purified from cells, performing the necessary experiments still took about 6 months. All in all, this project took us a year from start to finish, whereas testing everything purely experimentally would have taken us at least three times as long.”
Having obtained a Schmidt Science Fellowship, Roman will next undertake a postdoc in at the University of Washington in Seattle, delving much further into AlphaFold and protein structure prediction. “My time at TU Delft and with DelftBlue has been a fantastic preparation for this next step in my career. And I hope the administrators of the supercomputer over there will be just as open-minded and helpful.”