Biodiversity Assessment in Rivers Through eDNA Lab
THE PROBLEM
Biodiversity in river systems has significantly declined over the past decades. To address these declining trends and gain insights into the effectiveness of various conservation and restoration practices, there is a need for high-throughput biomonitoring methods. The detection of environmental DNA (eDNA) offers a promising technique to meet these needs. In addition to monitoring, it is necessary to develop modeling techniques that, through the analysis of hydrodynamics and morphodynamic changes, can predict habitat suitability for species.
WHAT WE DO
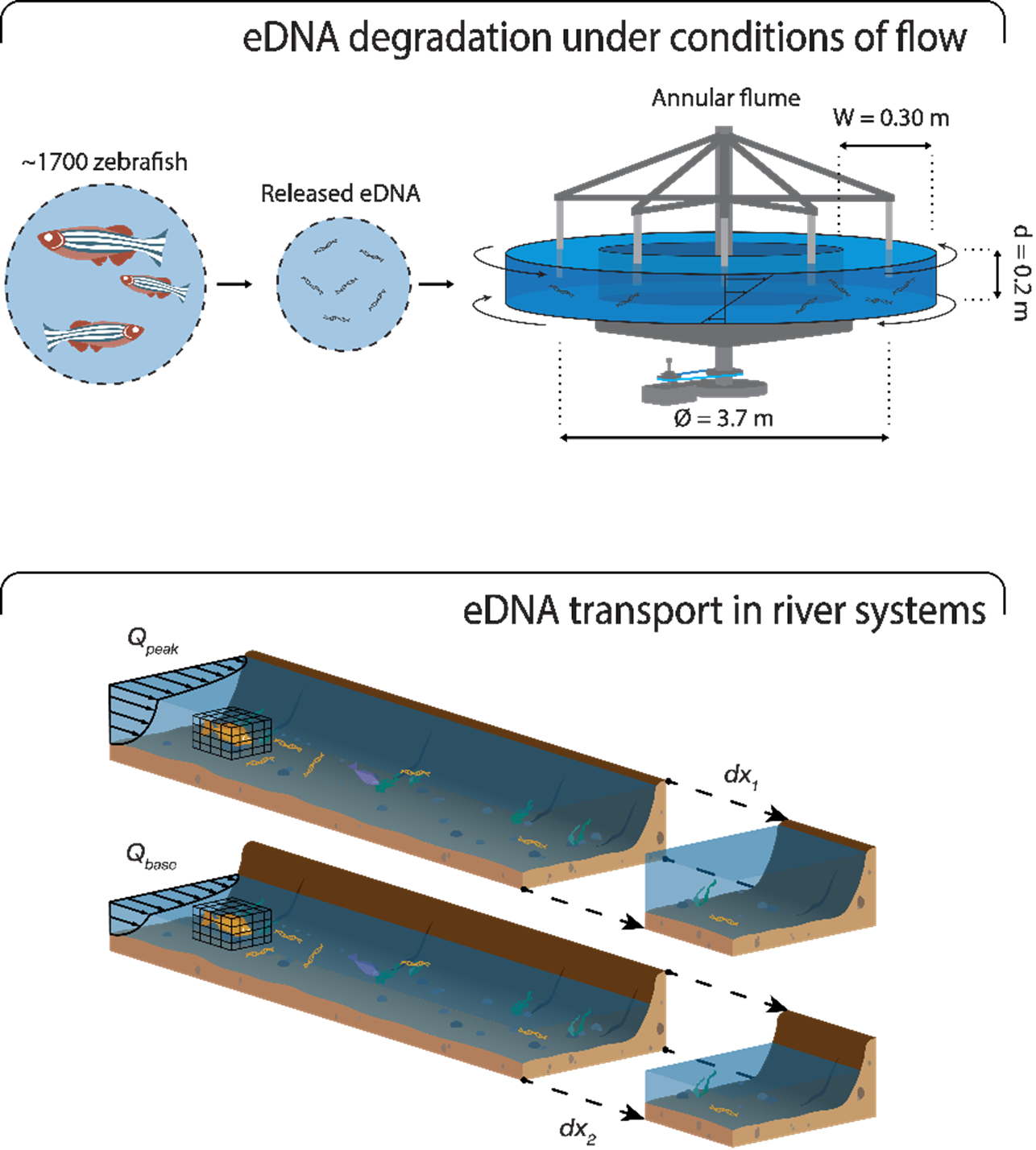
In our research team we employ a multi-disciplinary approach to uncover eDNA transport dynamics in river systems using a combination of novel genetic analysis techniques and hydraulic modelling. In doing so we aim to reduce uncertainties currently associated with the application of eDNA-based techniques in river systems. Our most recent research is centered on evaluating the degradation of environmental DNA (eDNA) under varying flow conditions and assessing the longitudinal transport of genetic material in rivers.
CURIOUS TO KNOW MORE ABOUT eDNA?
Organisms produce and release DNA in their direct surroundings over their lifespan. After release, the DNA degrades through time, while being transported through the environment either as free-floating material, or as encapsulated material in the form of cells, tissues, gametes, and organelles. By sampling and analysing this eDNA, community compositions of local biodiversity can then be inferred. In comparison to classical sampling approaches, eDNA-based monitoring is non-lethal for most taxonomic groups, reduces habitat disruption, and uses a singular standardized sampling protocol that can be applied for biodiversity across the tree of life.
ONGOING PROJECTS
- PhD research project Jelle Dercksen
- DECODE - bioDivErsity estimate for aquatiC ecOsystems aDopting eDNA (funded by: Topconsortium Kennis en Innovatie (TKI) Deltatechnologie, PI: Laura M. Stancanelli): it aims to advance the application of eDNA-based strategies for monitoring aquatic biodiversity by investigating how genetic material is dispersed in water. The research activities are carried out by a consortium that links expertise in species care (senior aquarist, Diergaarde Blijdorp), genetics (senior scientist, Naturalis Biodiversity Center), environmental science (senior scientists, Leiden University and Radboud University), and hydraulic transport (senior scientist, Delft University of Technology). In addition, we collaborate with key stakeholders in the user sector such as Deltares, Rijkswaterstaat, and Waardenburg Ecology.
MAIN CONTACTS
THE PROBLEM
Biodiversity in river systems has significantly declined over the past decades. To address these declining trends and gain insights into the effectiveness of various conservation and restoration practices, there is a need for high-throughput biomonitoring methods. The detection of environmental DNA (eDNA) offers a promising technique to meet these needs. In addition to monitoring, it is necessary to develop modeling techniques that, through the analysis of hydrodynamics and morphodynamic changes, can predict habitat suitability for species.
WHAT WE DO
In our research team we employ a multi-disciplinary approach to uncover eDNA transport dynamics in river systems using a combination of novel genetic analysis techniques and hydraulic modelling. In doing so we aim to reduce uncertainties currently associated with the application of eDNA-based techniques in river systems. Our most recent research is centered on evaluating the degradation of environmental DNA (eDNA) under varying flow conditions and assessing the longitudinal transport of genetic material in rivers.
CURIOUS TO KNOW MORE ABOUT eDNA?
Organisms produce and release DNA in their direct surroundings over their lifespan. After release, the DNA degrades through time, while being transported through the environment either as free-floating material, or as encapsulated material in the form of cells, tissues, gametes, and organelles. By sampling and analysing this eDNA, community compositions of local biodiversity can then be inferred. In comparison to classical sampling approaches, eDNA-based monitoring is non-lethal for most taxonomic groups, reduces habitat disruption, and uses a singular standardized sampling protocol that can be applied for biodiversity across the tree of life.
ONGOING PROJECTS
- PhD research project Jelle Dercksen
- DECODE - bioDivErsity estimate for aquatiC ecOsystems aDopting eDNA (funded by: Topconsortium Kennis en Innovatie (TKI) Deltatechnologie, PI: Laura M. Stancanelli): it aims to advance the application of eDNA-based strategies for monitoring aquatic biodiversity by investigating how genetic material is dispersed in water. The research activities are carried out by a consortium that links expertise in species care (senior aquarist, Diergaarde Blijdorp), genetics (senior scientist, Naturalis Biodiversity Center), environmental science (senior scientists, Leiden University and Radboud University), and hydraulic transport (senior scientist, Delft University of Technology). In addition, we collaborate with key stakeholders in the user sector such as Deltares, Rijkswaterstaat, and Waardenburg Ecology.