Building a DNA segregation system for a synthetic cell
Beatriz Orozco Monroy and Reza Amini Hounejani
In this project, our team aims at building a DNA segregation system based on bacterial cytoskeletal filaments recruiting a cell-free gene expression mechanism. In this regard, we test different existing options including bacterial microtubules, TubZRC segregation system, ParMRC partitioning system, and PhuZ fialments. A third possible option for each of these systems is to combine any of the two systems to build a hybrid segregation machine which is controllable by light.
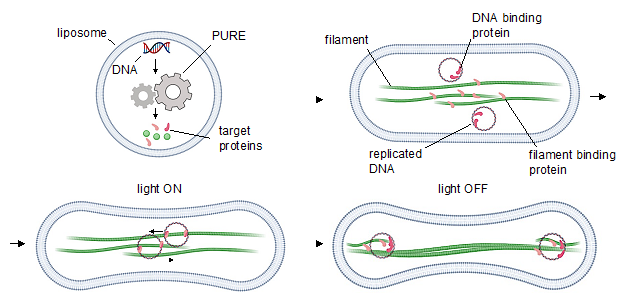
One way of dealing with the overwhelming complexity of cells, is to use reductionist approaches to gain a sufficient level of understanding of cellular phenomena. This can be achieved via two routes: top-down and bottom-up, each of which has its own advantages and disadvantages. Top-down approaches rely on removing the components of a system one by one and study the resulting effect on the system, whereas bottom-up approaches consist of the reconstitution of a specific cellular system or process using minimal components. While reconstitutions of complex systems are challenging and success has been limited, they provide an excellent tool to dissect the role of individual components under highly controlled conditions. One of the long-investigated mysteries of biology is the exact mechanism of cell division and DNA segregation and associated regulation pathways. So far minimal systems have been reconstituted that mimic prokaryotic DNA segregation but to date there has been no reconstitution of DNA segregation mediated by a bipolar eukaryotic mitotic spindle.