Publications
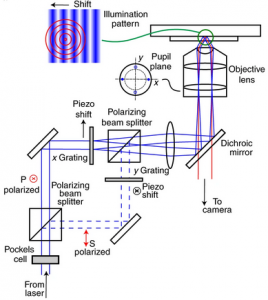
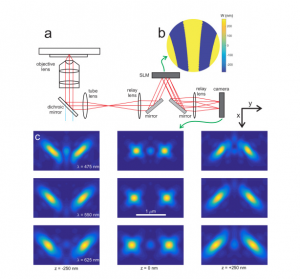
Localization microscopy at doubled precision with patterned illumination
MINFLUX offers a breakthrough in single molecule localization precision, but is limited in field of view. Here we combine centroid estimation and illumination pattern induced photon count variations in a conventional widefield imaging setup to extract position information over a typical micrometer-sized field of view. We show a near two-fold improvement in precision over standard localization with the same photon count on DNA-origami nanostructures and tubulin in cells, using DNA-PAINT and STORM imaging.
NATURE METHODS (2020)
DOI: 10.1038/S41592-019-0657-7
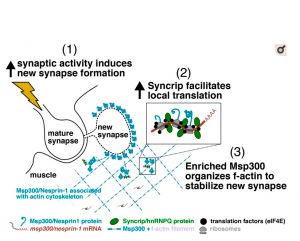
Syncrip/hnRNP Q is required for activity-induced Msp300/Nesprin-1 expression and new synapse formation.
Memory and learning involve activity-driven expression of proteins and cytoskeletal reorganization at new synapses, requiring posttranscriptional regulation of localized mRNA a long distance from corresponding nuclei. A key factor expressed early in synapse formation is Msp300/Nesprin-1, which organizes actin filaments around the new synapse. How Msp300 expression is regulated during synaptic plasticity is poorly understood. Here, we show that activity-dependent accumulation of Msp300 in the postsynaptic compartment of the Drosophila larval neuromuscular junction is regulated by the conserved RNA binding protein Syncrip/hnRNP Q. Syncrip (Syp) binds to msp300 transcripts and is essential for plasticity. Single-molecule imaging shows that msp300 is associated with Syp in vivo and forms ribosome-rich granules that contain the translation factor eIF4E. Elevated neural activity alters the dynamics of Syp and the number of msp300:Syp:eIF4E RNP granules at the synapse, suggesting that these particles facilitate translation. These results introduce Syp as an important early acting activity-dependent regulator of a plasticity gene that is strongly associated with human ataxias.
JOURNAL OF CELL BIOLOGY (2020)
DOI: 10.1083/JCB.201903135
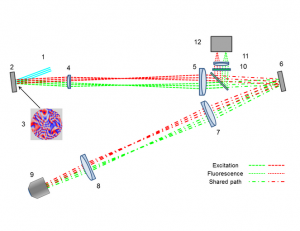
Anisoplanatic adaptive optics in parallelized laser scanning microscopy
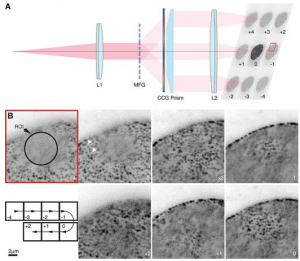
Inhomogeneities in the refractive index of a biological sample can introduce phase aberrations in microscopy systems, severely impairing the quality of images. Adaptive optics can be employed to correct for phase aberrations and improve image quality. However, conventional adaptive optics can only correct a single phase aberration for the whole field of view (isoplanatic correction) while, due to the three dimensional nature of biological tissues, the sample induced aberrations in microscopy often vary throughout the field of view (anisoplanatic aberration), limiting significantly the effectiveness of adaptive optics. This paper reports on a new approach for aberration correction in laser scanning confocal microscopy, in which a spatial light modulator is used to generate multiple excitation points in the sample to simultaneously scan different portions of the field of view with completely independent correction, achieving anisoplanatic compensation of sample induced aberrations. The method was tested in 150μm thick whole Drosophila brains, showing a dramatic improvement in resolution and sharpness when compared to conventional isoplanatic adaptive optics.
OPTICS EXPRESS (2020)
DOI: 10.1364/OE.389974
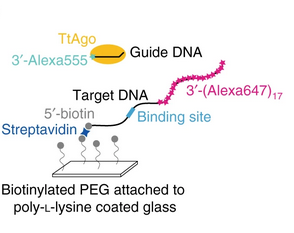
An automated Bayesian pipeline for rapid analysis of single-molecule binding data
Single-molecule binding assays enable the study of how molecular machines assemble and function. Current algorithms can identify and locate individual molecules, but require tedious manual validation of each spot. Moreover, no solution for high-throughput analysis of single-molecule binding data exists. Here, we describe an automated pipeline to analyze single-molecule data over a wide range of experimental conditions. In addition, our method enables state estimation on multivariate Gaussian signals. We validate our approach using simulated data, and benchmark the pipeline by measuring the binding properties of the well-studied, DNA-guided DNA endonuclease, TtAgo, an Argonaute protein from the Eubacterium Thermus thermophilus. We also use the pipeline to extend our understanding of TtAgo by measuring the protein’s binding kinetics at physiological temperatures and for target DNAs containing multiple, adjacent binding sites.
NATURE COMMUNICATIONS (2019)
DOI: 10.1038/S41467-018-08045-5
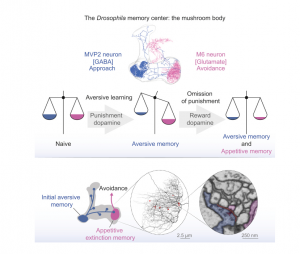
Integration of Parallel Opposing Memories Underlies Memory Extinction
Accurately predicting an outcome requires that animals learn supporting and conflicting evidence from sequential experience. In mammals and invertebrates, learned fear responses can be suppressed by experiencing predictive cues without punishment, a process called memory extinction. Here, we show that extinction of aversive memories in Drosophila requires specific dopaminergic neurons, which indicate that omission of punishment is remembered as a positive experience. Functional imaging revealed co-existence of intracellular calcium traces in different places in the mushroom body output neuron network for both the original aversive memory and a new appetitive extinction memory. Light and ultrastructural anatomy are consistent with parallel competing memories being combined within mushroom body output neurons that direct avoidance. Indeed, extinction-evoked plasticity in a pair of these neurons neutralizes the potentiated odor response imposed in the network by aversive learning. Therefore, flies track the accuracy of learned expectations by accumulating and integrating memories of conflicting events.
CELL (2018)
DOI: 10.1016/J.CELL.2018.08.021
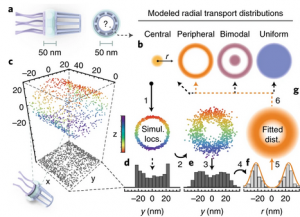
Deconstructing transport-distribution reconstruction in the nuclear-pore complex
Nuclear-pore complexes (NPCs) span the nuclear envelope and mediate bidirectional transport between the nucleus and cytoplasm. Macromolecules (>40 kDa) require transport receptors to transit the NPC efficiently, whereas smaller molecules diffuse through the NPC passively1. The mechanism through which the semipermeable barrier at the center of the NPC regulates selective transport is unknown. Aiming to elucidate this mechanism, Yang and colleagues have investigated spatial cargo distribution within the NPC2 by using single-point edge-excitation subdiffraction (SPEED) microscopy [...]
NATURE STRUCTURAL & MOLECULAR BIOLOGY (2018)
DOI: 10.1038/S41594-018-0161-2
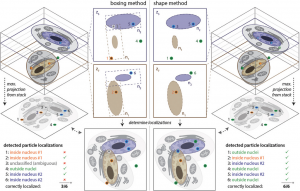
Advanced 3D Analysis and Optimization of Single-Molecule FISH in Drosophila Muscle
Single‐molecule fluorescence in situ hybridization (smFISH) provides direct access to the spatial relationship between nucleic acids and specific subcellular locations. The ability to precisely localize a messenger RNA can reveal key information about its regulation. Although smFISH is well established in cell culture or thin sections, the utility of smFISH is hindered in thick tissue sections due to the poor probe penetration of fixed tissue, the inaccessibility of target mRNAs for probe hybridization, high background fluorescence, spherical aberration along the optical axis, and the lack of methods for image segmentation of organelles. Studying mRNA localization in 50 µm thick Drosophila larval muscle sections, these obstacles are overcome using sample‐specific optimization of smFISH, particle identification based on maximum likelihood testing, and 3D multiple‐organelle segmentation. The latter allows independent thresholds to be assigned to different regions of interest within an image stack. This approach therefore facilitates accurate measurement of mRNA location in thick tissues.
SMALL METHODS (2018)
DOI: 10.1002/SMTD.201700324
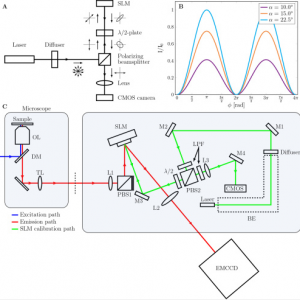
High precision wavefront control in point spread function engineering for single emitter localization
Point spread function (PSF) engineering is used in single emitter localization to measure the emitter position in 3D and possibly other parameters such as the emission color or dipole orientation as well. Advanced PSF models such as spline fits to experimental PSFs or the vectorial PSF model can be used in the corresponding localization algorithms in order to model the intricate spot shape and deformations correctly. The complexity of the optical architecture and fit model makes PSF engineering approaches particularly sensitive to optical aberrations. Here, we present a calibration and alignment protocol for fluorescence microscopes equipped with a spatial light modulator (SLM) with the goal of establishing a wavefront error well below the diffraction limit for optimum application of complex engineered PSFs. We achieve high-precision wavefront control, to a level below 20 mλ wavefront aberration over a 30 minute time window after the calibration procedure, using a separate light path for calibrating the pixel-to-pixel variations of the SLM, and alignment of the SLM with respect to the optical axis and Fourier plane within 3 μm (x/y) and 100 μm (z) error. Aberrations are retrieved from a fit of the vectorial PSF model to a bead z-stack and compensated with a residual wavefront error comparable to the error of the SLM calibration step. This well-calibrated and corrected setup makes it possible to create complex ‘3D+λ’ PSFs that fit very well to the vectorial PSF model. Proof-of-principle bead experiments show precisions below 10 nm in x, y, and λ, and below 20 nm in z over an axial range of 1 μm with 2000 signal photons and 12 background photons.
OPTICS EXPRESS (2018)
DOI: 10.1364/OE.26.008397
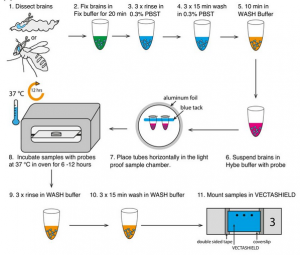
Single molecule fluorescence in situ hybridisation for quantitating post-transcriptional regulation in Drosophila brains
RNA in situ hybridization can be a powerful method to investigate post-transcriptional regulation, but analysis of intracellular mRNA distributions in thick, complex tissues like the brain poses significant challenges. Here, we describe the application of single-molecule fluorescent in situ hybridization (smFISH) to quantitate primary transcription and post-transcriptional regulation in whole-mount Drosophila larval and adult brains. Combining immunofluorescence and smFISH probes for different regions of a single gene, i.e., exons, 3’UTR, and introns, we show examples of a gene that is regulated post-transcriptionally and one that is regulated at the level of transcription. We also show that the method can be used to co-visualise a variety of different transcripts and proteins in neuronal stems cells as well as deep brain structures such as mushroom body neuropils. Finally, we introduce the use of smFISH as asensitivealternative to conventional antibody labelling to mark specific neural stem cell populations in the brain.
METHODS (2017)
DOI: 10.1016/J.YMETH.2017.06.025
Simultaneous measurement of emission color and 3D position of single molecules
We show that the position of single molecules in all three spatial dimensions can be estimated alongside its emission color by diffractive optics based design of the Point Spread Function (PSF). The phase in a plane conjugate to the aperture stop of the objective lens is modified by a diffractive structure that splits the spot on the camera into closely spaced diffraction orders. The distance between and the size of these sub-spots are a measure of the emission color. Estimation of the axial position is enabled by imprinting aberrations such as astigmatism and defocus onto the orders. The overall spot shape is fitted with a fully vectorial PSF model. Proof-of-principle experiments on quantum dots indicate that a spectral precision of 10 to 20 nm, an axial localization precision of 25 to 50 nm, and a lateral localization precision of 10 to 30 nm can be achieved over a 1 μm range of axial positions for on average 800 signal photons and 17 background photons/pixel. The method appears to be rather sensitive to PSF model errors such as aberrations, giving in particular rise to biases in the fitted wavelength of up to 15 nm.
OPTICS EXPRESS (2016)
DOI: 10.1364/OE.24.004996
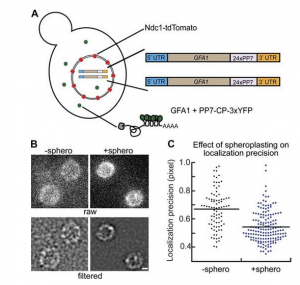
In vivo single-particle imaging of nuclear mRNA export in budding yeast demonstrates an essential role for Mex67p
Many messenger RNA export proteins have been identified; yet the spatial and temporal activities of these proteins and how they determine directionality of messenger ribonucleoprotein (mRNP) complex export from the nucleus remain largely undefined. Here, the bacteriophage PP7 RNA-labeling system was used in Saccharomyces cerevisiae to follow single-particle mRNP export events with high spatial precision and temporal resolution. These data reveal that mRNP export, consisting of nuclear docking, transport, and cytoplasmic release from a nuclear pore complex (NPC), is fast (∼200 ms) and that upon arrival in the cytoplasm, mRNPs are frequently confined near the nuclear envelope. Mex67p functions as the principal mRNP export receptor in budding yeast. In a mex67-5 mutant, delayed cytoplasmic release from NPCs and retrograde transport of mRNPs was observed. This proves an essential role for Mex67p in cytoplasmic mRNP release and directionality of transport.
JOURNALL OF CELL BIOLOGY (2015)
DOI: 10.1083/JCB.201503135
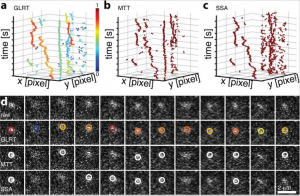
Probability-based particle detection that enables threshold-free and robust in vivo single-molecule tracking
Single-molecule detection in fluorescence nanoscopy has become a powerful tool in cell biology but can present vexing issues in image analysis, such as limited signal, unspecific background, empirically set thresholds, image filtering, and false-positive detection limiting overall detection efficiency. Here we present a framework in which expert knowledge and parameter tweaking are replaced with a probability-based hypothesis test. Our method delivers robust and threshold-free signal detection with a defined error estimate and improved detection of weaker signals. The probability value has consequences for downstream data analysis, such as weighing a series of detections and corresponding probabilities, Bayesian propagation of probability, or defining metrics in tracking applications. We show that the method outperforms all current approaches, yielding a detection efficiency of >70% and a false-positive detection rate of <5% under conditions down to 17 photons/pixel background and 180 photons/molecule signal, which is beneficial for any kind of photon-limited application. Examples include limited brightness and photostability, phototoxicity in live-cell single-molecule imaging, and use of new labels for nanoscopy. We present simulations, experimental data, and tracking of low-signal mRNAs in yeast cells.
MOLECULAR BIOLOGY OF THE CELL (2015)
DOI: 10.1091/MBC.E15-06-0448
A 4D view on mRNA
Imaging single molecules in live cells in 4+D (space, time and colors) is crucial for studying various biological processes, especially for observing the behavior of RNA molecules within the nuclear landscape [1]. RNA molecules are known to serve a multitude of tasks such as being templates for protein translation or to act as enzymes for regulating countless reactions in the nucleus [1]. Studying RNA kinetics in living cells can provide new information on RNA function or even human diseases, for instance caused by viruses such as the human immunodeficiency virus (HIV) [2]. A challenge to imaging nuclear RNA function is that the nucleus as a whole undergoes major reformation during the cell cycle [1] but the time required to step through the sample limits the capability to image large numbers of rapidly moving particles in a 3D space.
ONCOTARGET (2015)
DOI: 10.18632/ONCOTARGET.5121
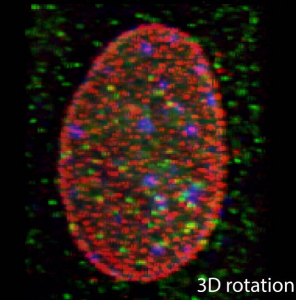
Nuclear accessibility of β-actin mRNA is measured by 3D single-molecule real-time tracking
Imaging single proteins or RNAs allows direct visualization of the inner workings of the cell. Typically, three-dimensional (3D) images are acquired by sequentially capturing a series of 2D sections. The time required to step through the sample often impedes imaging of large numbers of rapidly moving molecules. Here we applied multifocus microscopy (MFM) to instantaneously capture 3D single-molecule real-time images in live cells, visualizing cell nuclei at 10 volumes per second. We developed image analysis techniques to analyze messenger RNA (mRNA) diffusion in the entire volume of the nucleus. Combining MFM with precise registration between fluorescently labeled mRNA, nuclear pore complexes, and chromatin, we obtained globally optimal image alignment within 80-nm precision using transformation models. We show that β-actin mRNAs freely access the entire nucleus and fewer than 60% of mRNAs are more than 0.5 µm away from a nuclear pore, and we do so for the first time accounting for spatial inhomogeneity of nuclear organization.
JOURNAL OF CELL BIOLOGY (2015)
DOI: 10.1083/JCB.201411032
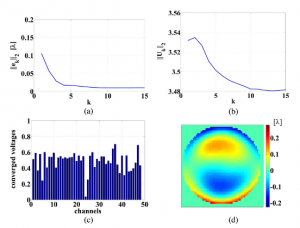
Iterative learning control of a membrane deformable mirror for optimal wavefront correction
We present an iterative learning control (ILC) algorithm for controlling the shape of a membrane deformable mirror (DM). We furthermore give a physical interpretation of the design parameters of the ILC algorithm. On the basis of this insight, we derive a simple tuning procedure for the ILC algorithm that, in practice, guarantees stable and fast convergence of the membrane to the desired shape. In order to demonstrate the performance of the algorithm, we have built an experimental setup that consists of a commercial membrane DM, a wavefront sensor, and a real-time controller. The experimental results show that, by using the ILC algorithm, we are able to achieve a relatively small error between the real and desired shape of the DM while at the same time we are able to control the saturation of the actuators. Moreover, we show that the ILC algorithm outperforms other control algorithms available in the literature.
APPLIED OPTICS (2013)
DOI: 10.1364/AO.52.002363
Iterative linear focal-plane wavefront correction
We propose an efficient approximation to the nonlinear phase diversity (PD) method for wavefront reconstruction and correction from intensity measurements with potential of being used in real-time applications. The new iterative linear phase diversity (ILPD) method assumes that the residual phase aberration is small and makes use of a first-order Taylor expansion of the point spread function (PSF), which allows for arbitrary (large) diversities in order to optimize the phase retrieval. For static disturbances, at each step, the residual phase aberration is estimated based on one defocused image by solving a linear least squares problem, and compensated for with a deformable mirror. Due to the fact that the linear approximation does not have to be updated with each correction step, the computational complexity of the method is reduced to that of a matrix-vector multiplication. The convergence of the ILPD correction steps has been investigated and numerically verified. The comparative study that we make demonstrates the improved performance in computational time with no decrease in accuracy with respect to existing methods that also linearize the PSF.
JOURNAL OF THE OPTICAL SOCIETY OF AMERICA A (2013)
DOI: 10.1364/JOSAA.30.002002
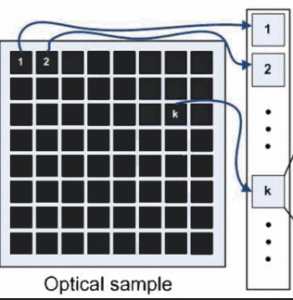
Focal-plane wavefront estimation and control using the extended Kalman filter
This paper addresses the problem of disturbance rejection for faint Poisson point-like measurements. The aberration function is approximated with a finite Zernike based expansion. We use nonlinear observers to estimate the aberrations and a linear quadratic regulator to reject the aberrations. Kalman filtering is compared with the phase diversity maximum aposteriori estimation. The approach presented here is beneficial for instance for 2-photon observations, single molecule observations or natural/laser guide star observations in astronomy.
IEEE (2012)
DOI: 10.1109/ISOT.2012.6403289
Fast, single-molecule localization that achieves theoretically minimum uncertainty
We describe an iterative algorithm that converges to the maximum likelihood estimate of the position and intensity of a single fluorophore. Our technique efficiently computes and achieves the Cramér-Rao lower bound, an essential tool for parameter estimation. An implementation of the algorithm on graphics processing unit hardware achieved more than 10(5) combined fits and Cramér-Rao lower bound calculations per second, enabling real-time data analysis for super-resolution imaging and other applications.