Molecular toolbox
Our group gives the opportunity to access the state of the art next generation cloning, genome editing technologies such as CRISPR. In the past four years we conducted in collaboration P. Daran-Lapujade group the development and the implementation of several key methodologies in the field of strain engineering that improved and accelerated the pace of the research in the industrial microbiology section.

Mans, R., van Rossum, H. M., Wijsman, M., Backx, A., Kuijpers, N. G., van den Broek, M., Daran-Lapujade, P., Pronk, J. T., van Maris, A. J. and Daran, J. M. (2015). FEMS Yeast Res 15. fov004.
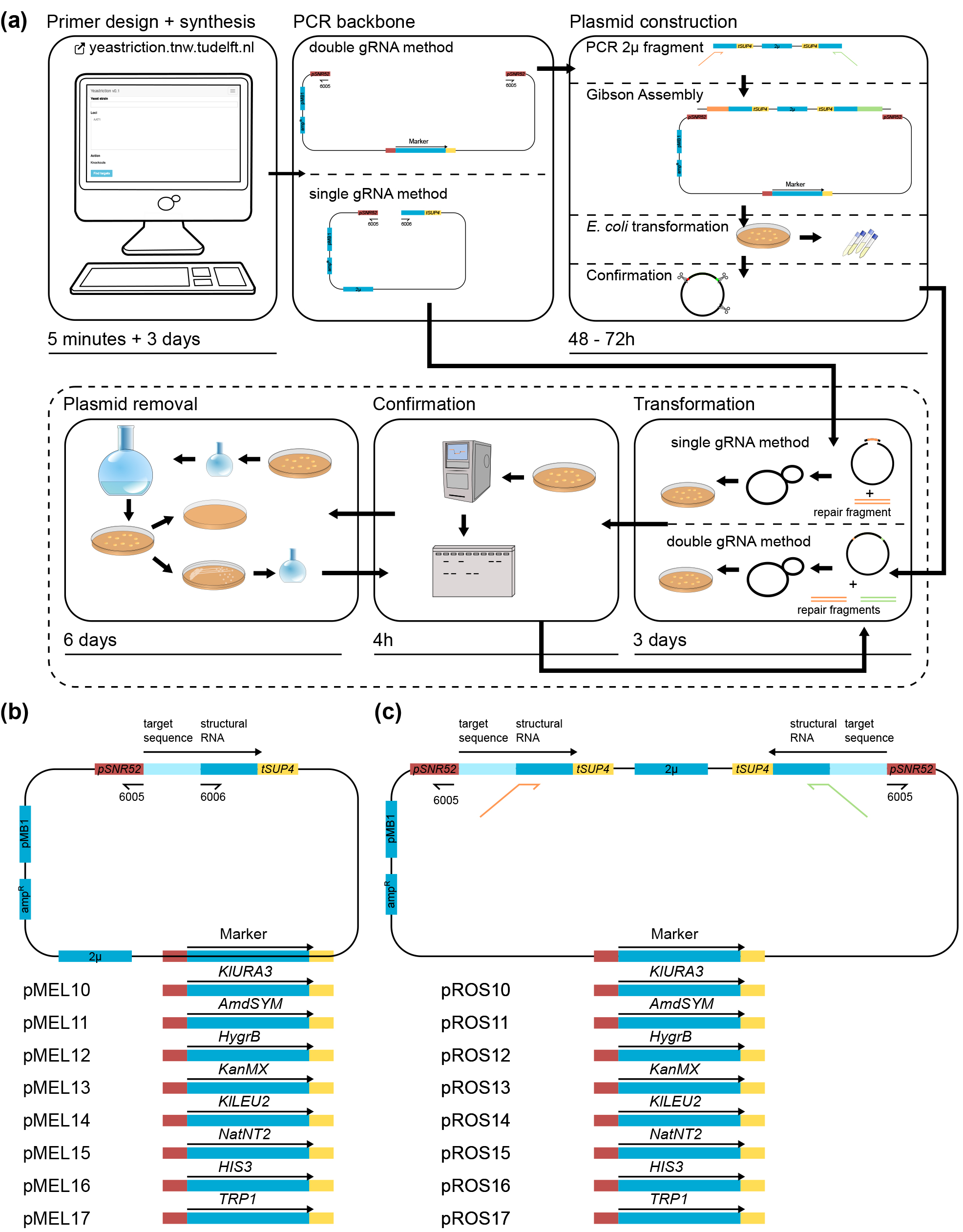
CRISPR/Cas9-based approaches enables fast(er) strain construction. They exploit the malleability of this molecular scissor to guide the simultaneous introduction of multiple genetic modifications. Sets of standardized plasmids, as well as the web-based Yeastriction target-sequence identifier and primer-design tool, are made available to the yeast research community to facilitate fast, standardized and efficient application of the CRISPR/Cas9 system.
Plasmids and strains are available at http://www.euroscarf.de/search.php?search=&project=Projects&gene=CRISPR%2FCas9&selectedProject=Projects&updateSelection=
Mans, R., van Rossum, H. M., Wijsman, M., Backx, A., Kuijpers, N. G., van den Broek, M., Daran-Lapujade, P., Pronk, J. T., van Maris, A. J. and Daran, J. M. (2015). FEMS Yeast Res 15. fov004.
CRISPR/Cas9-based approaches enables fast(er) strain construction. They exploit the malleability of this molecular scissor to guide the simultaneous introduction of multiple genetic modifications. Sets of standardized plasmids, as well as the web-based Yeastriction target-sequence identifier and primer-design tool, are made available to the yeast research community to facilitate fast, standardized and efficient application of the CRISPR/Cas9 system.
Plasmids and strains are available at http://www.euroscarf.de/search.php?search=&project=Projects&gene=CRISPR%2FCas9&selectedProject=Projects&updateSelection=

Recyclable markers, counter selectable cassettes that can be removed from the targeted genome after use, are therefore valuable assets in ambitious metabolic engineering programs., the new recyclable dominant marker cassette amdSYM, formed by the Ashbya gossypii TEF2 promoter and terminator and a codon-optimized acetamidase gene (Aspergillus nidulans amdS), has been constructed. The amdSYM cassette confers S. cerevisiae the ability to use acetamide as sole nitrogen source. Direct repeats flanking the amdS gene allow for its efficient recombinative excision. As previously demonstrated in filamentous fungi, loss of the amdS marker cassette from S. cerevisiae can be rapidly selected for by growth in the presence of fluoroacetamide. The amdSYM cassette can be used in different genetic backgrounds and represents the first counterselectable dominant marker gene cassette for use in S. cerevisiae. amdSYM is a useful addition to the genetic engineering toolbox for Saccharomyces laboratory, wild, and industrial strains.
Solis-Escalante, D., Kuijpers, N. G., Bongaerts, N., Bolat, I., Bosman, L., Pronk, J. T., Daran, J. M. and Daran-Lapujade, P. (2013).. FEMS Yeast Res 13, 126-39.
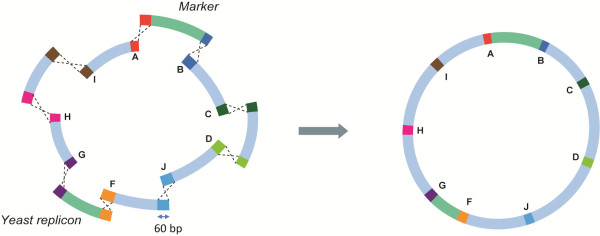
In vivo assembly of overlapping fragments by homologous recombination in Saccharomyces cerevisiae is a powerful method to engineer large DNA constructs. We explores the potential of combining in vivo assembly of large, multigene expression constructs with their targeted chromosomal integration in S. cerevisiae. I-SceI-assisted integration dramatically increased the efficiency of assembly and integration of the same construct to 95%. This study paves the way for the fast, efficient, and stable integration of large DNA constructs in S. cerevisiae chromosomes.
Kuijpers, N. G., Chroumpi, S., Vos, T., Solis-Escalante, D., Bosman, L., Pronk, J. T., Daran, J. M. and Daran-Lapujade, P. (2013a). FEMS Yeast Res 13, 769-81.
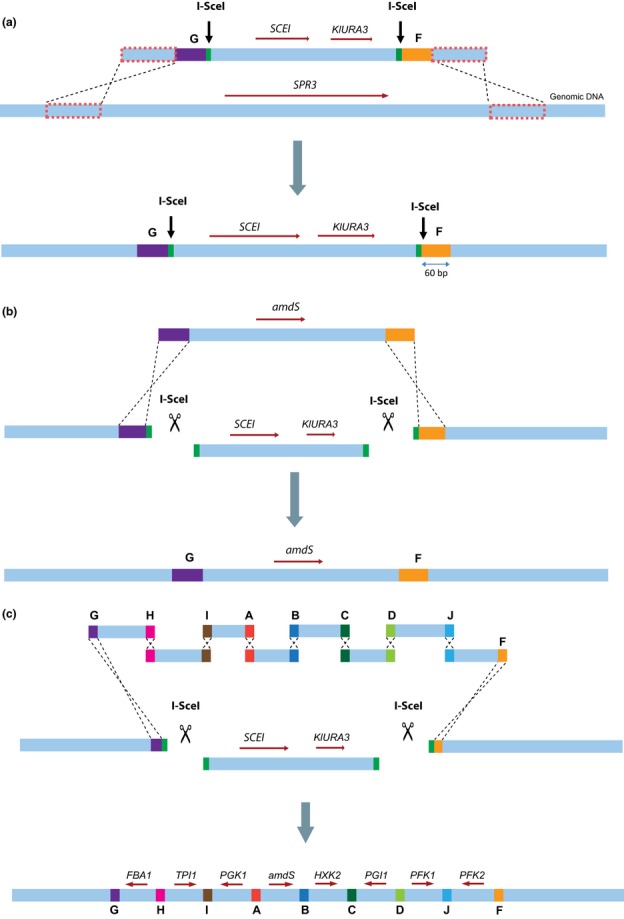
This method delivers a substantial improvement for assembly of multi-fragment expression vectors in S. cerevisiae. Not only does it improve the efficiency of in vivo assembly, but it also offers a versatile platform for easy and rapid design and assembly of synthetic constructs. The presented method is therefore ideally suited for the construction of complex pathways and for high throughput strain construction programs for metabolic engineering purposes. In addition its robustness and ease of use facilitate the construction of any plasmid carrying two or more genes.
Kuijpers, N. G., Solis-Escalante, D., Bosman, L., van den Broek, M., Pronk, J. T., Daran, J. M. and Daran-Lapujade, P. (2013b).. Microb Cell Fact 12, 47.